Get maps of spatially resolved properties
Once we finished SED fitting to all spatial bins in the galaxy, we can construct maps of properties from the collection of those fitting results (in FITS files). For this, we can use piXedfit.piXedfit_fitting.maps_parameters() function in the piXedfit_fitting module. Please see the API reference here for more information about this function. An example of script to get the maps properties is shown below.
from piXedfit.piXedfit_fitting import maps_parameters fits_binmap = "pixbin_corr_specphoto_fluxmap_ngc309.fits" hdu = fits.open(fits_binmap) nbins = int(hdu[0].header['NBINSPH']) hdu.close() bin_ids = [] name_sampler_fits = [] for ii in range(0,nbins): bin_ids.append(ii) name = "mcmc_bin%d.fits" % (ii+1) name_sampler_fits.append(name) fits_fluxmap = "corr_specphoto_fluxmap_ngc309.fits" name_out_fits = "maps_properties.fits" maps_parameters(fits_binmap=fits_binmap, bin_ids=bin_ids, name_sampler_fits=name_sampler_fits, fits_fluxmap=fits_fluxmap,refband_SFR=0, refband_SM=11, name_out_fits=name_out_fits)
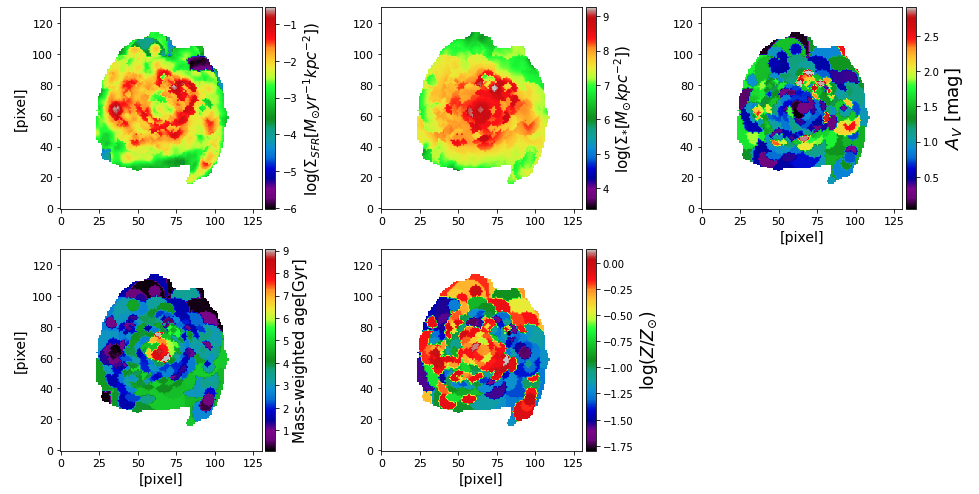
We can then make plots from the maps stored in the output FITS file.
maps = fits.open("maps_properties.fits") gal_region = maps['galaxy_region'].data # calculate physical size of a single pixel from piXedfit.piXedfit_images import kpc_per_pixel z = float(maps[0].header['gal_z']) arcsec_per_pixel = 1.5 # pixel size in arcsec kpc_per_pix = kpc_per_pixel(z,arcsec_per_pixel) rows, cols = np.where(gal_region == 0) # get pixels outside the galaxy's region of interest # stellar mass surface density map_prop_SM = np.log10(np.power(10.0,maps['pix-log_mass-p50'].data)/kpc_per_pix/kpc_per_pix) map_prop_SM[rows,cols] = float('nan') # surface density of SFR map_SFR = np.log10(np.power(10.0,maps['pix-log_sfr-p50'].data)/kpc_per_pix/kpc_per_pix) map_SFR[rows,cols] = float('nan') # Av dust attenuation map_Av = 1.086*maps['bin-dust2-p50'].data map_Av[rows,cols] = float('nan') # stellar metallicity map_logzsol = maps['bin-logzsol-p50'].data map_logzsol[rows,cols] = float('nan') # Mass-weighted age map_mw_age = np.power(10.0,maps['bin-log_mw_age-p50'].data) map_mw_age[rows,cols] = float('nan') # plotting from mpl_toolkits.axes_grid1 import make_axes_locatable fig1 = plt.figure(figsize=(14,7)) ###=> SFR surface density f1 = fig1.add_subplot(2, 3, 1) plt.ylabel('[pixel]', fontsize=14) plt.setp(f1.get_xticklabels(), fontsize=11) plt.setp(f1.get_yticklabels(), fontsize=11) plt.imshow(map_SFR, origin='lower', cmap='nipy_spectral') ax = plt.gca() divider = make_axes_locatable(ax) cax = divider.append_axes("right", size="5%", pad=0.05) cb = plt.colorbar(cax=cax) cb.set_label(r'log($\Sigma_{SFR}[M_{\odot}yr^{-1}kpc^{-2}]$)', fontsize=15) ###=> stellar mass surface density f1 = fig1.add_subplot(2, 3, 2) plt.setp(f1.get_xticklabels(), fontsize=11) plt.setp(f1.get_yticklabels(), fontsize=11) plt.imshow(map_prop_SM, origin='lower', cmap='nipy_spectral') ax = plt.gca() divider = make_axes_locatable(ax) cax = divider.append_axes("right", size="5%", pad=0.05) cb = plt.colorbar(cax=cax) cb.set_label(r'log($\Sigma_{*}[M_{\odot}kpc^{-2}]$)', fontsize=15) ###=> AV dust attenuation f1 = fig1.add_subplot(2, 3, 3) plt.xlabel('[pixel]', fontsize=14) plt.setp(f1.get_xticklabels(), fontsize=11) plt.setp(f1.get_yticklabels(), fontsize=11) plt.imshow(map_Av, origin='lower', cmap='nipy_spectral') ax = plt.gca() divider = make_axes_locatable(ax) cax = divider.append_axes("right", size="5%", pad=0.05) cb = plt.colorbar(cax=cax) cb.set_label(r'$A_{V}$ [mag]', fontsize=18) ### mass-weighted age f1 = fig1.add_subplot(2, 3, 4) plt.xlabel('[pixel]', fontsize=14) plt.ylabel('[pixel]', fontsize=14) plt.setp(f1.get_xticklabels(), fontsize=11) plt.setp(f1.get_yticklabels(), fontsize=11) plt.imshow(map_mw_age, origin='lower', cmap='nipy_spectral') ax = plt.gca() divider = make_axes_locatable(ax) cax = divider.append_axes("right", size="5%", pad=0.05) cb = plt.colorbar(cax=cax) cb.set_label('Mass-weighted age[Gyr]', fontsize=15) ### stellar metallicity f1 = fig1.add_subplot(2, 3, 5) plt.xlabel('[pixel]', fontsize=14) plt.setp(f1.get_xticklabels(), fontsize=11) plt.setp(f1.get_yticklabels(), fontsize=11) plt.imshow(map_logzsol, origin='lower', cmap='nipy_spectral') ax = plt.gca() divider = make_axes_locatable(ax) cax = divider.append_axes("right", size="5%", pad=0.05) cb = plt.colorbar(cax=cax) cb.set_label(r'log($Z/Z_{\odot}$)', fontsize=17) plt.subplots_adjust(left=0.07, right=0.95, bottom=0.1, top=0.98, hspace=0.2, wspace=0.3) plt.show()
We can then get a plot as shown below.